
5'-AMP-activated protein kinase catalytic subunit alpha-1
The identification of binding pockets in proteins is crucial for understanding protein function, drug discovery, annotating protein function, predicting protein-protein interactions, and facilitating structural studies. GENEOnet provides a foundation for advancing the knowledge of biological systems and developing new therapeutic strategies.

5'-AMP-activated protein kinase catalytic subunit alpha-1
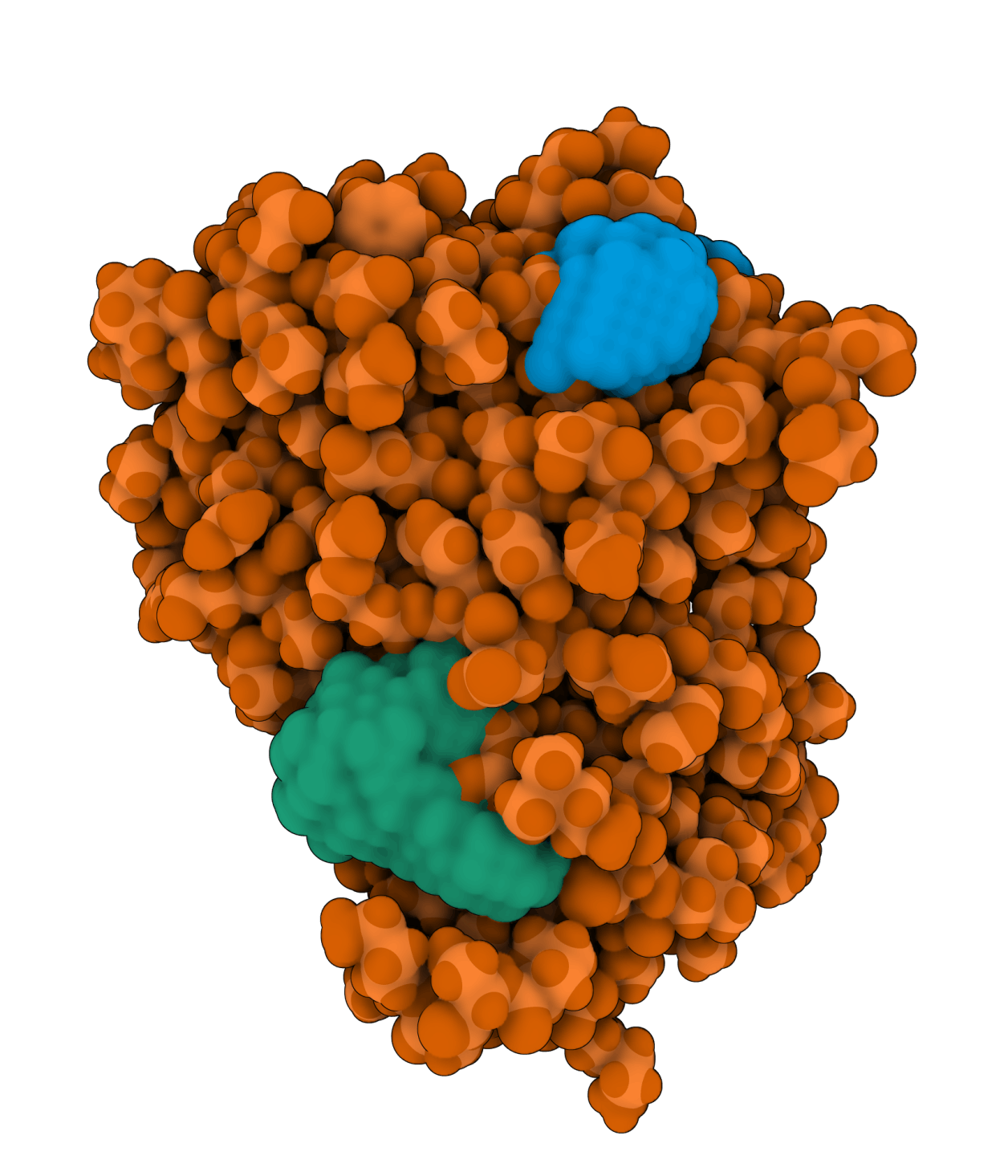
Aurora kinase A
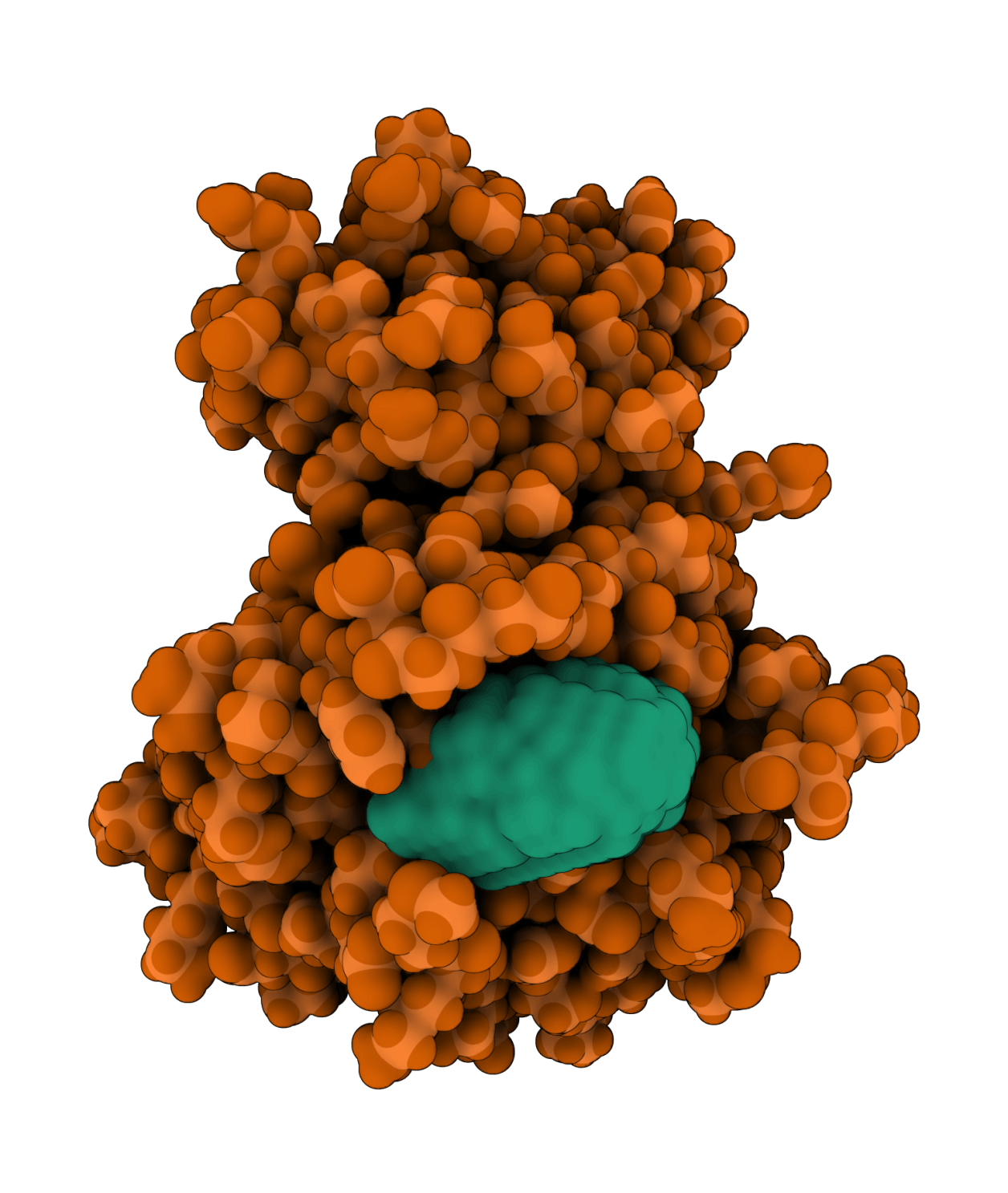
5'-AMP-activated protein kinase catalytic subunit alpha-1